| Molecular
biology has progressed at an amazing rate in the last
two decades yielding a set
of tools
that allow
us to manipulate DNA in a very controlled way. The aim
of this section of the courses is to show a set of examples
of the
different tools that can be used to solve a wide variety
of problems. Our claim
is that, at least when dealing with E. coli, it is mostly
about asking the right question rather than developing new techniques.
The particular goal
of the project is to extract a gene from the E. coli genome
(lacZ) and insert it into a plasmid that will control that
gene under
a tetracycline inducible promoter (pZE21, Lutz and Bujard,
1997). In order to accomplish this the students have to become
acquainted
with restricition enzymes, gel
electrophoresis, PCR, ligation,
transformations, sequencing and
measuring gene expression at the single cell level using GFP.
Besides the obvious
objective of manipulating DNA we had an extra question in
mind. As an introduction to measuring gene expression we raised
the
question: "does the relative change in gene expression
depend on the reporter gene that is used?". The relevance
of this question is obvious if one is trying to be quantitative...
the message should not depend on the messenger! This particular question is one of the advanced projects that some students take place in towards the end of the bootcamp.
Careful
planning and understanding of the involved processes and
their results is needed. We used Invitrogen's
VectorNTI software
in an intensive way, which allowed us to graphically understand
what would happen to the DNA molecules in each step of
this series of experiments. During these types of tutorials all students get to use the software on their computers and plan the next steps for the particular DNA molecule they will do cloning on.
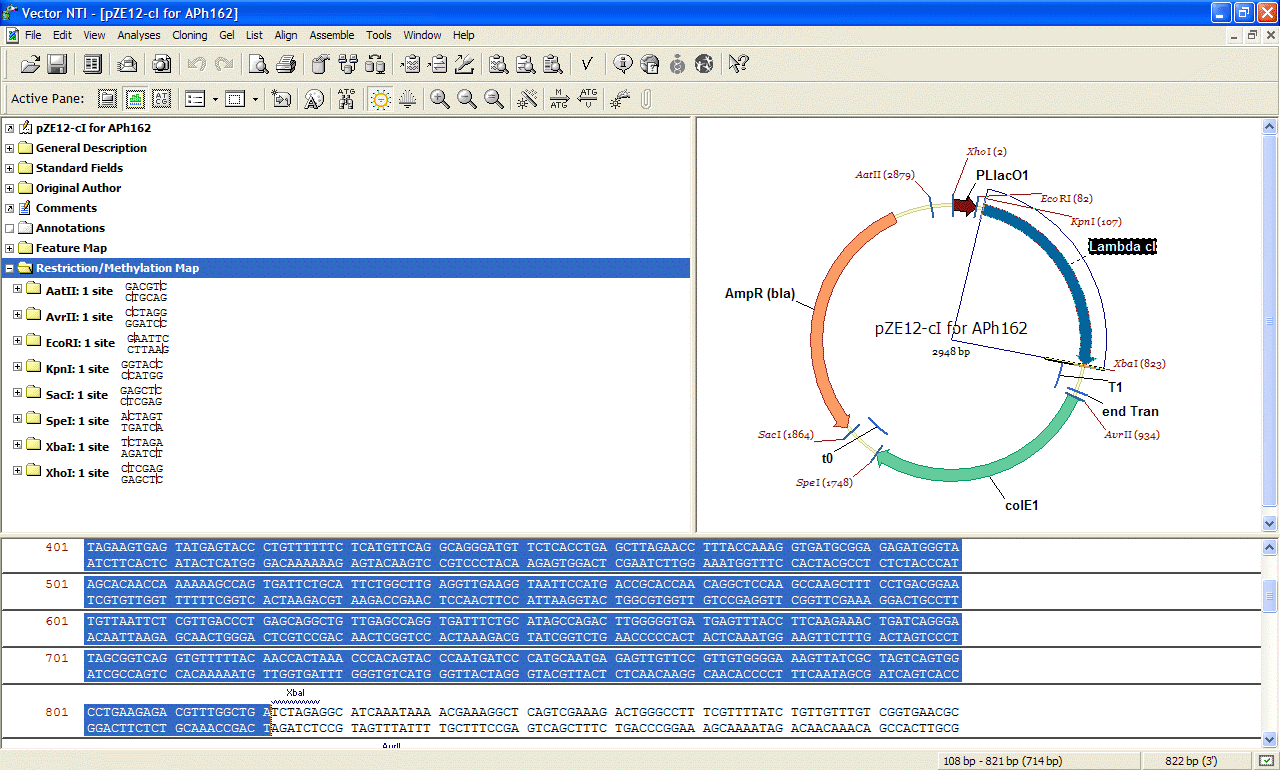 |
Vector NTI
allows to view commented DNA, and its features making
it easy
to predict restriction fragments, design
PCR or sequencing primers and do BLAST searches of
particular features of the DNA sequence we might be
interested in.
|
General Concept We used the pZ system
developed by Lutz and Bujard as our vector. Its main advantage
is that it has unique restriction sites around each one
of its important features (coding region, antibiotic resistance,
origin
of replication, etc.), making it a highly modular plasmid
system. 
We
worked with pZE21-GFP which expresses GFP under the control
of a tetracycline inducible promoter (PLtetO-1). The plasmid
also has resistance against the antibiotic kanamycin and its
origin of replication is ColE1 (50-70 copies per cell).
Restriction
Digest and Gel Electrophoresis The
original vector was digested with the restriction enzymes
KpnI and HindIII and the samples were run on an agarose gel.
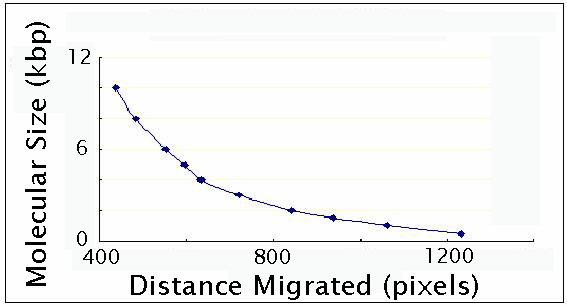
This
is a good chance for the students to perform a gel calibration
using a DNA ladder.
Two
bands are visible in the double digest, one corresponding to
the vector (~2200bp) and the other one corresponding to the
GFP gene (~700bp), which we discard..
Using
Qiagen's Gel
Extraction Kit we purified the band corresponding to the
vector (pZE21), which was going to be ligated with the lacZ
insert.
|
|
| 
