Artificial Pattern Formation in E.ColiDavid Van Valen Oren Schaedel Michael Amori |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Plasmid Design Materials and Methods Results |
Abstract
Weiss et al created a band pass filter using genetically engineered building blocks and used them to create spatial patterns of gene expression. We attempted to reconstruct this experiment in order to investigate effects of AHL diffusion on the generation of spatial patterns as a function of distance, location, and timing. To reconstruct this experiment, we first transformed DH5a cells with plasmids received from the Weiss group. We then set out to perform two sets of experiments on our transformed cells. First, we wished to characterize the band pass filter by measuring the expression of GFP as a function of AHL concentration. Second, we wished to examine the types of spatial patterns we could generate by varying the location of AHL emitting cells. Unfortunately our experiments were impeded by a number of technical dilemmas and we were only able to verify the band-pass nature of one plasmid strand. Plasmid Design
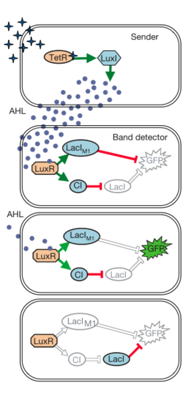
The design of pHD and pLD allows the concomitant transcription of CI and LacIm1 by the transcriptional regulator LuxR (which is activated by AHL). LacIm1 inhibits the Lac promoter on which GFP is present and CI represses the lambda promoter, onto which the LacI is transcribed. Thus at high AHL concentrations, both CI and LacIm1 are transcribed shutting off GFP expression. CI inhibition is stronger than LacIm1 shutting off the native LacI, which represses GFP, therefore at medium AHL concentrations, LacIm1 is less effective in pLac inhibition and GFP is turned on. At low AHL concentrations, CI is transcribed at basal levels, allowing transcription of LacI (WT) which represses GFP transcription. When describing this as a function of distance, sender cells emit AHL which diffuses as a gradient throughout the plate. High concentrations are close to the sender cells, and low concentrations are present far form the cells. Plating cells with both pLD and pHD around sender cells forms a fluorescent ring due to the gradient of AHL. 3 different versions of pHD were constructed, high sensitivity (HD3), medium sensitivity (HD2) and low sensitivity (HD1). HD3 cells have a low copy number of LuxR, therefore higher AHL concentrations are required to successfully create the high detect filter. HD2 cells have a WT LuxR, and HD1 cells have a hypersensitive mutant of LuxR, activated by lower concentrations of AHL. Figure 1 Design of a band-pass filter Materials and Methods
We developed a number of protocols that helped streamline our experiments Transformation
Cell Culture Pipette 5 mL of LB into a falcon tube. The antibiotics added depends on the strain that use used. No antibiotics are added for wildtype cultures, chloramphenicol for the single plasmid HD, and chloramphenicol plus kanamycin for the double strain LD. 15uL of chloramphenicol and 10uL od kanamycin are added per 5mL of media. Once the culture media is prepared, we inoculate the culture with cells from our frozen stocks. The cells are grown in the 37 ˇC room overnight Induction The cells for the induction experiment should be placed in
cell culture the previous night. We always prepare the wildtype cells in
addition to the transformed cells because measurements of their fluorescence
are needed to calculate the fold change. 30 uL of the cultured cells are
placed in 8mL of M9 media with the appropriate antibiotics several hours
before the planned experiment because M9 produces less auto-fluorescence. The
experiment also required a solution of 100mM AHL dissolved in phosphate
buffer.
To achieve these concentrations, we first performed a 1:10
dilution of the 100mM AHL stock. It should be noted that if we wished to
explore different orders of magnitude, all we have to do is either perform a
higher dilution of the stock, or just use the stock directly. We then
pipetted 220 uL of cells into row A and 180 uL into rows B-D. The following
volumes of the 10 mM AHL solution were added:
Once the AHL is added, we then performed a serial 1:10
dilution of row A. 20 uL from row A are pipetted into row B, 20 uL of row B
are pipetted into row C, and 20 uL from row B are pipetted into row D. The
space between the wells is filled with dH2O and the plate is
covered with the special plastic top generously provided by Dhananjay Tampe
to prevent evaporation during culture. The plate is then cultured overnight
in the plate reader. The proper setup file is accessible on snowdome, as are
the MATLAB files we created to interpret the data.
, where all values are taken relative the values of the M9 media. Cell Plating This experiment was orchestrated in order to test whether
the band pass filter functioned as a function of distance from a point
source. Cells emitting AHL were plated in the center of the petri dish, and
BD or HD cells were plated around. A double layer of M9-agar was plated in
the following fashion: the bottom layer consisted of M9-agar at 1.5% with
appropriate antibiotics was poured and let to dry. A second layer of M9-agar
0.7% was plated on top premixed with the receiving cells, and chloramphenicol
only (sender cells have only Chloramphenicol resistance). After second layer
dried, filter paper was cut at the diameter of a 1 ml tube, loaded with
sender cells and placed in the center of the petri dish. Two plates were made
per strain. Controls for this setup were a WT dish, and a sender dish W/O
receiver cells. Results
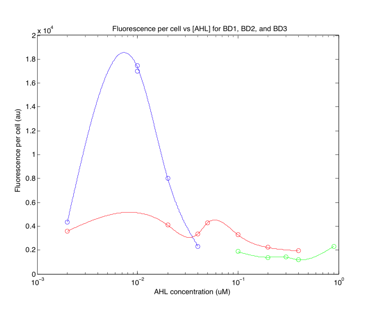
Figure 2 Fluorescence response of
transformed cells to AHL Unfortunately, we only had one successful trial of the induction experiment. While we were unable to calculate fold-change of gene expression in this experiment, the fluorescence per cell clearly shows a band pass behavior for the BD1 plasmid. From our first experiment, we thought that the concentration points we used were simply to spread out to properly observe the peak of BD2, and did not include high enough concentrations to observe the peak for BD3. We aimed to repeat the experiment for the BD2 and BD3 strains using more a better range of concentrations. Unfortunately, we ran into a problem during the next set of experiments. We expect our cells to be in the log phase of growth, but when we ran the experiment again, we observed a constant OD600, implying no growth. Plotting the OD600 as a function of AHL concentration, we saw the following for BD2:
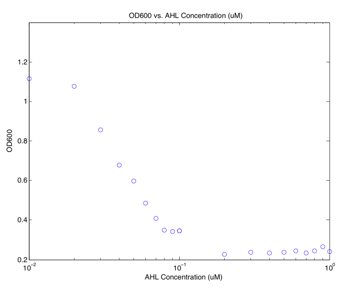
Figure 3 OD600 as a function of AHL
concentration for BD2 From the graph, we can see that the OD600 decreases as the concentration of AHL increases? Why is that? After some investigation, we were able to determine that the acidic ethyl acetate that the AHL was dissolved in was actually dissolving polystyrene. Almost everything that we used in the lab was made of polystyrene, including the plates used for the plate reader. We attempted to redo the experiment using lower concentrations of AHL, and hence less solvent but we were unsuccessful. Our solution was to order AHL as a solid compound and dissolve it in a phosphate buffer to make a 100mM solution. Working with AHL in a different buffer solves the ethyl acetate problem, but unfortunately we had issues with cell growth in the M9 media. As a result, none of our most recent experiments can provide any useful data. In parallel with our efforts to characterize the plasmids, we also plated them to see if we would get the pattern formation described by Weiss et al. The experiment was rather lengthy, and we were only able to attempt it once. We did not see any fluorescence on the plates. Our main suspicion is that either the cells were unable to grow in the media, or that the production of AHL and its diffusion through the agar was so high and so fast that we missed the window where the concentration in the plate was within the necessary concentration range. Conclusions
Since we donŐt have many results, we can advise of steps to improve future attempts.
We had several experiments which came to mind:
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||