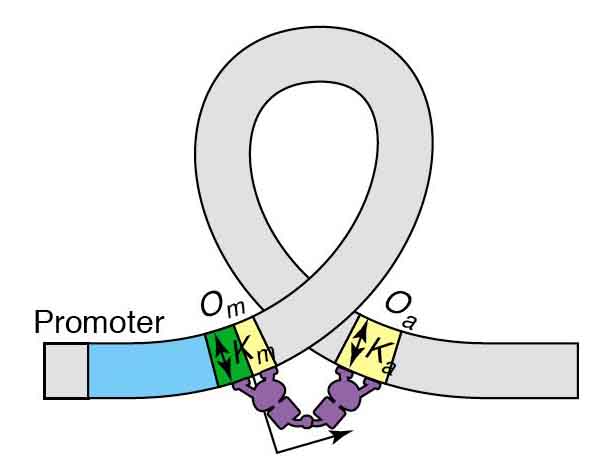
A pZS22 plasmid has been successfully modified to express YFP under the controll of a recombinant LacUV5 promoter with a single O1 operator site immediately upstream of the -30 RNA binding site. This construct will be used in future analyses of the impact of DNA sequence on looping and, in turn, LacI repression efficiency at the promoter. Initial measurements indicate that induction levels for the O1-LacUV5 promoter construct will be on the same order as those observed for pZS22-YFP plasmids. The Lac operon of E. coli discovered by Jacob Monod in the late 1940's has been studied in such detail that it has ascended to the status of a hydrogen atom in the realm of gene regulation. The basic principles behind the operation of the Lac operon are well established and are explained in depth at a number of resources. The biophysical properties of the lac operon are being investigated to transcend a phenomological or cartoon model understanding of this system. For example, Müller-Hill recently investigated the influence of distance between operator sites in the lac operon on repression by LacI and discovered a periodicity in repression efficiency of around 10 bp, which is the number of base pairs in a full turn of the DNA helix. This implies that twisting of DNA is energetically unfavorable for repression by LacI (see Figure 1, Müller-Hill 1996). This study points toward a role of DNA mechanics in repression by LacI; a phenomenon which may have general implications for a number of known repression mechanisms involving looping.  Figure 1: The role of DNA mechanics in gene regulation at the Lac operon (Müller, 1996). Keeping with the spirit of understanding the relationship between DNA mechanics and efficiency of repression, Bintu et al. (2005) have developed a statistical mechanics model of the Lac operon. This model includes looping -- which is known to play a prominent role in the proper operation of the lac operon -- as parameter by accounting for the energy required to loop the DNA between two Lac operator sites bound by LacI as shown in Figure 2 below, where Om and Oa are operator sites and the purple shape is the LacI protein dimer. To both verify the validity of the model and to be able to calculate a key parameter necessary for its use, it is necessary to quantify and fully understand the relationship between the mechanical properties of DNA between, and perhaps also around, the operator sites shown in Figure 2 and the ability of LacI to repress gene expression.
Cloutier and Widom (2004 and 2005) have recently reported work analyzing the relationship between looping potential and DNA sequence in vitro. This is a key step toward beginning to understand the biophysical phenomenon influencing gene regulation as relates to DNA looping. Unfortunately though, this work was done in vitro, far removed from the natural biological context of DNA looping. The work described herein describes the initial steps being taken by the Rob Phillips group to measure, in vitro, the role of DNA sequence on looping potential and, in turn, efficiency of repression at the Lac operon. The goal is to vary the sequence between the Om and Oa sites shown in Figure 2 -- note that the natural Lac operon has 3 sites, but can be readily manipulated to have only 2 -- and evaluate the impact on repression. This quantitative data may then be used to gain the insights alluded to above. This in vitro measurement is limited at present though because the promoter proximal operator in the naturally occuring Lac operon is downstream of the operator, therby restricting part of the sequence between the two operators to maintain a functional promoter. A simple solution to this problem is to move the operator upsptream of the promoter and hope that repression is not altered significantly. The construction of this modified promoter sequence and the verification that it works properly are reported herin. This work was completed as part of rotation project in the laboratory of Professor Rob Phillips at the California Insitute of Technology. |