|
The primers described in methods ordered from IDT were annealed as described. To evaluate the efficiency of the annealing process, a 1 ul sample of a 10X dilution of each oligomer solution was run on a 4% agarose gel. The gels were run for 1 to 1.5 hr in the abscence of any dye and were stained for 3 hours in 10 ug/mL SYBR gold stain. The gel was run in SB buffer and not the typical TAE buffer to lower the temperature of the gel during the run -- SB has a lower conductivity than TAE. The image of the gel obtained is shown below as Figure 1. The efficiency of annealing was approximately 50%. The uppermost band in each of the two banded wells represents the oligomer of interest. Double stranded DNA and single stranded DNA were resolved successfully on this gel. As a control, one lane contained a small sample from one of the primers (a O1-LacUV5 primer) used in the annealing reactions. The resolution of the gel is poor as a consequence of diffusion. The leftmost lane in the image below is the marker (NEB's low moleclar weight DNA ladder). 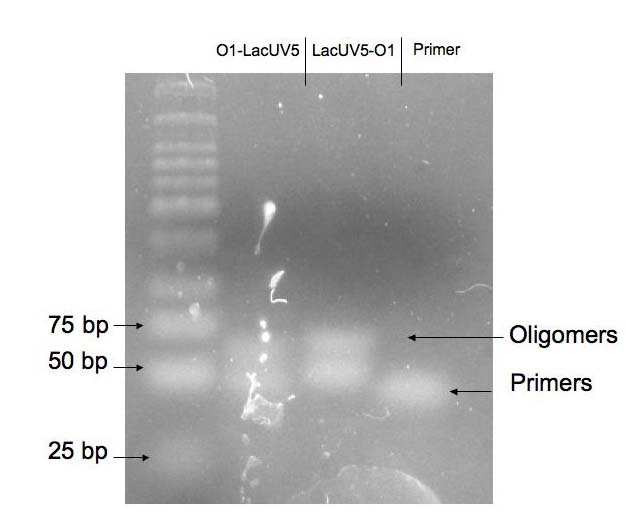 Figure 1: Image of gel run to evaluate efficiency of primer annealing. Back to Top. Double Digest and Purification of pZS22-YFP pZS22-YFP plasmids were extracted from cells using a mini prep kit and double digested as described. Prior to extracting the band of interest from the gel, an image was taken of the gel to verify that the plasmid was fully digested and of the expected length. An image of this gel is shown below as Figure 2. An undigested plasmid was run as a control and the marker used was NEB's 1 kb DNA ladder. As expected, the digest product had a molecular weight of around 4 kb and the undigested plasmid showed two bands corresponding to two states of a covalently linked circular plasmid.
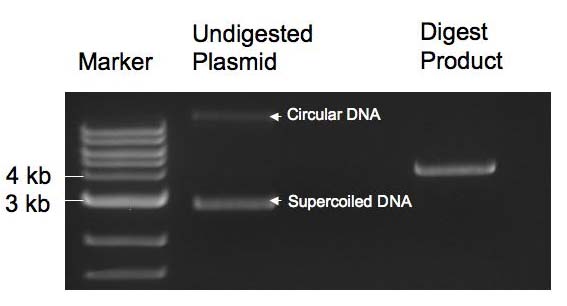 Figure 2: Image of gel from which double digested pZS22-YFP was extracted. Ligation of Plasmid and Transformation of Cells The plasmid and oligomers were ligated together as described and electoporation was then used to transform MC4100Z1 cells with the construct. The plasmids were transformed into this strain because it overexpresses LacI and it was anticipated that induction of YFP expression by IPTG would be particularly high and measurable in this strain. No growth was observed on day two of the experiment, so the entirety of the remaining transformed cell suspensions from day one were plated onto the agar plates and growth was then observed on the next day. The images in Figure 3 below was taken of the four transformants on this third day.
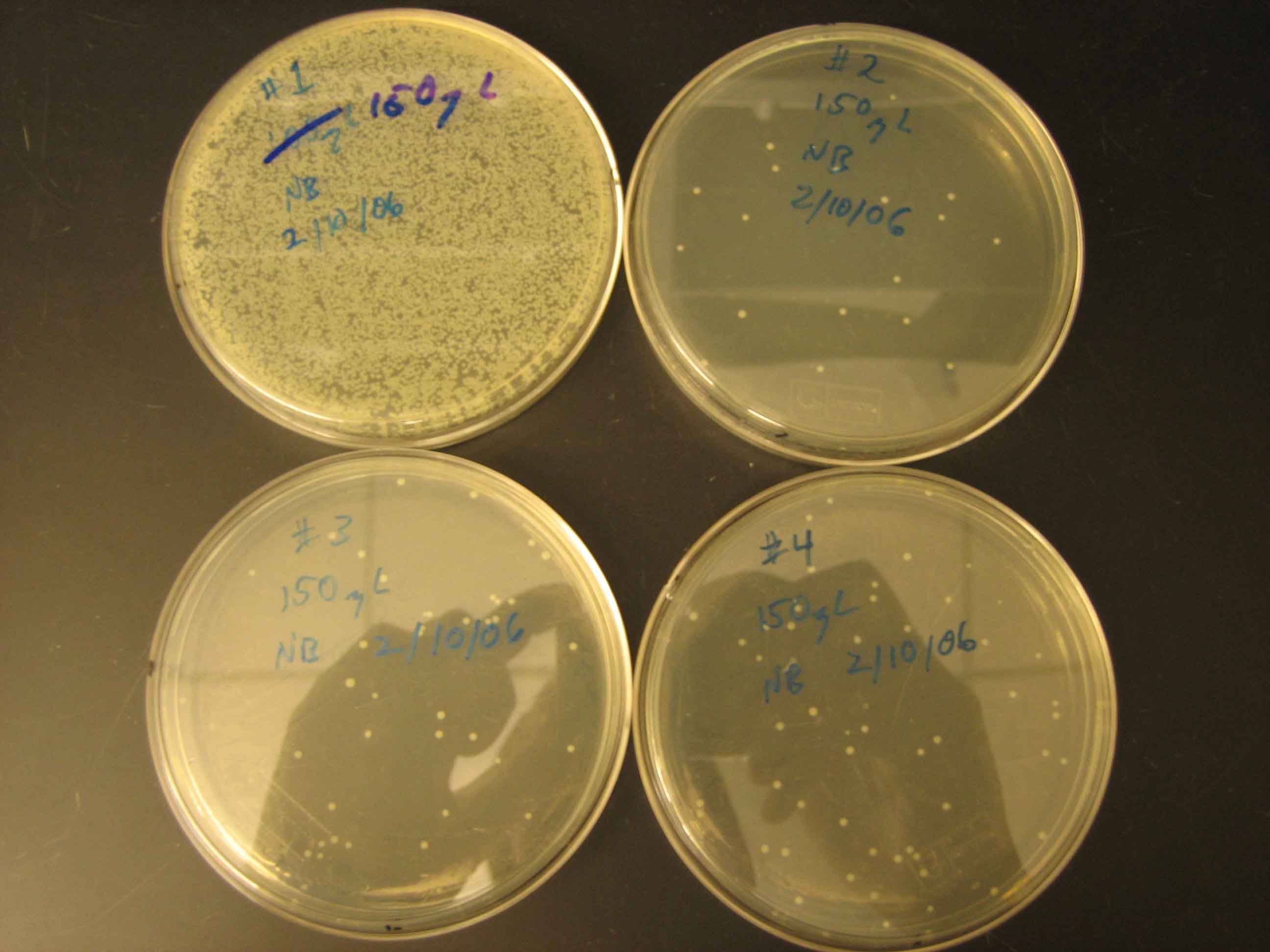 Figure 4: Transformmation Results It can be seen in Figure 4 that the efficiency of transformation was quite low for three of the samples. This can be avoided in the future by using heat shock of chemically compotent cells in the future, which is a technique with a higher transformation efficiency (pg DNA will yield transformants successfully). The plates in the above image were labeled as follows:
Back to Top. Three colonies were selected from each of the plates shown in Figure 4 and used to inoculate 5 ml LB medium supplemented with Kan antibiotic. From these cultures (12 in total), glycerol stocks were created and plasmids were extracted for sequencing using a mini prep kit (see methods). The sequencing results indicated that two of the cultures (ie. two of the plasmids sequenced) were very similar in sequence to the desired construct sequences. A midi prep was then used to prepare a large amount of the two plasmids of interest. These plasmids, when sequenced, were exact matches for the two plasmid constructs desired. The two promoter constructs had, therefore, been successfully integrated into pZS plasmids. The two strains harboring these plasmids were used in all future work. Preparation of Cells Harboring Constructs and Control Plasmids The midi preps had such a high concentration of DNA that only a fraction of the solutions had been submitted for sequencing. 1 ul of the remaining solutions were used to transform MG1655 and MG1655 ΔLacI (see methods). MG1655 ΔLacI does not express LacI, so it will consitutively express YFP when transformed with the pZS constructs. Comparing expression levels of YFP in MG1655 and MG1655 ΔLacI allowed for the measurement of induction relative to basal levels of YFP expression in a wild type strain. Additionally, as a control, both strains were transformed with the origonal pZS22-YFP plasmid used to create the constructs. Glycerol stocks were prepared using isolated colonies on LB + Kan agar for each of the above transformations. Bulk Fluorescence Measurements Fluorescence was measured in bulk as described on the methods page for YFP under control of each of the promoter constructs, created as described above, and for a pZS22-YFP control. The bar graph below summarizes the results of these measurements. Each culture was evaluated in triplicate. Each yellow bar represents the ratio of YFP expression in the LacI delete strain to that in the LacI expressing strain. Each YFP measurement was normalized to optical density. The error bars in Figure 5 represent one standard deviation.
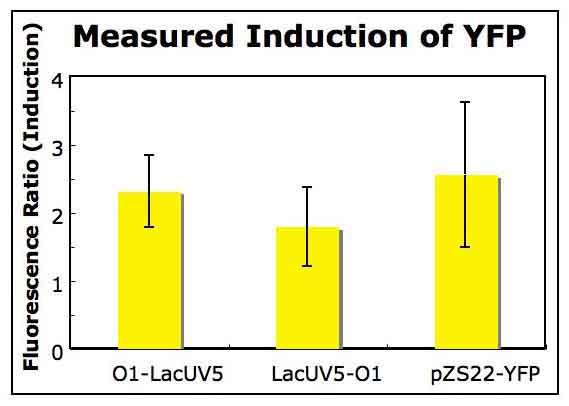 Figure 5: Measuring induction of each promoter construct using a bulk fluorescence measurement. The O1-LacUV5 construct was observed to effectively repress expression of YFP. This repression may be investigated further using single-cell measurements or flow cytometry, which will allow for the measurement of gene expression in single cells. The efficient repression of gene expression by the O1-LacUV5 construct does not come as any great surprise (Guido, 2006), but it is a necessary verification to begin investigating the phenomena discussed in the introduction. Unfortunately, the autofluorescence of negative control cells not harboring the YFP expressing plasmids was too high to allow for its subtraction from the basal level of YFP expression in MG1655 cells. For this reason, the induction levels in Figure 5 are artificially low -- perhaps by a few orders of magnitude. This limitation of the experiment may be overcome in the future by investigating the autofluorescence of these strains further. |